Original Article

Survivin Expression and Correlation with Clinico-pathological Parameters in Breast Cancer
Kumkum Jha1, Mohan Kumar2, Vijay K Shukla3, Manoj Pandey1
- 1Department of Surgical Oncology, Institute of Medical Sciences, Banaras Hindu University, Varanasi 221 005, India
- 2Department of Pathology, Institute of Medical Sciences, Banaras Hindu University, Varanasi 221 005, India
- 3Department of General Surgery,Institute of Medical Sciences, Banaras Hindu University, Varanasi 221 005, India
- Submitted: February 7, 2012;
- Accepted February 27, 2012
- Published: March 10, 2012
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction:
Survivin is an intracellular, multifunctional protein which controls cell proliferation, inhibition of apoptosis and the promotion of angiogenesis. Present study deals with expression of survivin in fibroadenoma and breast cancer and correlation of its expression with other clinico-pathological parameters.
Materials and Methods:
Survivin expression was measured by Immunohistochemistry, Western Blotting and RT PCR in 29 cases of fibroadenoma and 91 breast cancer .
Results:
Survivin mRNA was present in 44.8% of fibroadenoma and 72.5% of breast cancer cases while survivin protein was expressed in 41.3% of fibroadenoma and 63.7% of breast cancer. Survivin expression showed significant correlation with tumor Stage, histological Grade, estrogen receptor (ER), and HER2 status .
Conclusion:
Increased survivin expression at gene level might serve as a diagnostic and prognostic marker in breast cancer and can be used as a therapeutic target.
Introduction
Breast cancer is influenced by the underlying hormonal milieu in which these cancers arise [1,2].Normal breast development is controlled by a balance between cell proliferation and apoptosis, and there is also a strong evidence that tumor growth is not just a result of uncontrolled proliferation but also of reduced apoptosis. Tumor cells can acquire resistance to apoptosis by expression of certain anti – apoptopic proteins called Inhibitors of Apoptosis Proteins (IAP). These IAPs were first discovered in baculoviruses, where they were shown to be involved in suppressing host cell death response to viral infection [3,4].Human genome encodes eight IAP family members including X-linked inhibitor of apoptosis protein (X-IAP), cIAP1, cIAP2, ML-IAP (Livin; K-IAP), Naip, ILP2 (TS-IAP), pollon/Bruce and survivin [5].
Survivin is an intracellular, multifunctional protein which controls cell proliferation [6], inhibition of apoptosis [6] and the promotion of angiogenesis [6]. Survivin is located on chromosome 11E2, contains three separate and chemically distinct surfaces which include acidic and basic regions on the BIR domain and a hydrophobic helical surface on alpha 6. Survivin is one of the chromosome passenger proteins and plays an important role in mitosis and spindle check points. It is also one of the top “transcriptomes” which are expressed in cancer but not in normal tissues with the exception of testis, thymus and placenta. Survivin is overexpressed in almost all cancers including lung, colon, breast, pancreas, stomach, liver, ovary and prostate cancer, as well as melanoma and hematopoietic malignancies [7-9]. Survivin shuttles between nucleus and the cytoplasm and hence can associate with different subcellular components. Survivin present in cytoplasmic compartment of cell is involved in initiation of cancer cell proliferation while the nuclear survivin leads to longer cell survival. For breast cancer patients, its prognostic value has been reported to be nonexistent [10-12], associated with improved outcome [13], or associated with adverse outcome [6,14,15]. Despite these conflicting findings, survivin is one of the 16 cancer-related genes represented in the Oncotype DXTM assay [16]. In the present study we have observed survivin expression at mRNA level and at protein level by using specific primers and antibodies.
Materials and method
Patients
After obtaining a written informed consent samples were obtained from 91 patients of breast cancer and 29 benign breast diseases (fibroadenoma) attending the Surgical Oncology and General Surgery outpatient, and scheduled to undergo surgery as part of their treatment. The study was approved by the institute ethics committee. After collection, the samples were snapped freezed in liquid nitrogen and were stored at -84 °C till further processing. A portion of the sample was stored in buffered formalin for histology and immunochemistry.
Immunohistochemistry
Four micron sections were prepared on poly L-lysin coated glass slides from the formalin fixed paraffin embedded tissues. Sections were transferred to xylene with three changes of 5min each. Rehydration was done with graded alcohol from 100% to 70% followed by running tap water for 30 min. Antigen retrieval was done in citrate buffer for 20min.Slides were washed 3 times after retrieval in TBS for 10 minutes each. Endogenous peroxidase activity was blocked by incubating section with 0.3% peroxide (H202) in methanol for 20 minutes. Sections were washed in TBS for 5 minutes and were incubated in normal blocking serum for 1 hour to block non-specific binding sites. Slides were incubated overnight with Survivin antibody (Survivin(D8):SC-17779, Santa Cruz Biotechnology, Inc.) Slides were washed in TBS for 5 minutes. Sections were incubated with secondary antibody (Vectastain Universal Elite Abc Kit). Slides were washed for 5 minutes in TBS. Sections were incubated in peroxidase substrate solution for color development. Finally sections were counterstained with hematoxylin for observation under light microscope for image analysis and quantification.
Grading of immunostaining
Immunostaining intensity was graded as Strong(+++),Moderate(++),Weak(+) or nil(0) staining.
RT-PCR
Total RNA was extracted by TRIZOL (Invitrogen) and reverse transcribed to cDNA with High Capacity cDNA Reverse Transcription Kits (Applied Biosystem). PCR reactions were performed using GAPDH as an internal standard. Primers used were: survivin forward: 5’-AAG AAC TGG CCC TTC TTG GA-3’, survivin reverse: 5’-CAA CCG GAC GAA TGC TTT T-3’.The predicted length of the amplified product was 185 bp [17]; GAPDH Forward Primer(FP): 5’-ACGGATTTGGTCGTATTGGGCG-3’,GAPDH Reverse Primer(RP) 5’-CTCCTGGAAGATGGTGATGG-3’.The predicted length of that amplified product was 212 bp [18]. The PCR cycles were set for total 33 cycles with denaturation temperature at 94 ° C for 30 s, annealing temperature at 54 ° C for 40 sec and extending temperature at 72 ° C for 1 min per cycle and last cycle lasted for 10 min at 72 ° C.
Western Blotting
Tissue were diluted in sodium dodecyl sulphate (SDS) buffer containing 50mM Tris-HCL (pH 6.8), 4% beta mercaptoethanol, 4% SDS, 20%
Table 1: Survivin Expression
| Analysis Method |
Fibroadenoma |
Breast Cancer |
p value |
| IHC |
41.3%(12/29) |
63.7%(58/91) |
0.064ns |
| Western Blotting |
40%(8/20) |
64.8% (46/71) |
0.005 |
| RT PCR |
44.8%(13/29) |
44.8%(13/29) |
0.079ns |
glycerol and 0.01% bromophenol blue. Protein from each sample was separated on a 15% SDS–polyacrylamide gel. Following electrophoresis, the separated proteins were transferred to a PVDF transfer membrane (Thermoscientific) using a wet blotting apparatus for overnight. The membranes were then treated for 1 hour at room temperature with a blocking solution containing 50mM Tris buffered saline (TBS), 5% skimmed milk powder and 0.05%Triton-X-100 (TBS-T). Following this, the membranes were incubated overnight at 4ºC with Survivin antibody (Survivin(D8):SC-17779, Santa Cruz Biotechnology, Inc.), in a blocking solution containing 50mM TBS-T and 5% skimmed milk powder. Subsequently, membranes were washed in 50mM TBS containing 0.05% Triton and then probed with Goat anti mouse IgG-HRP (GeNei) secondary antibody for 4 hrs. The blots were washed three times for 10 min each in 50mM TBS-T. Immunoreactivity was detected using DAB reagent (GeNei). As a control for equal loading, membranes were reprobed with mouse anti-β -actin antibody (Sigma).
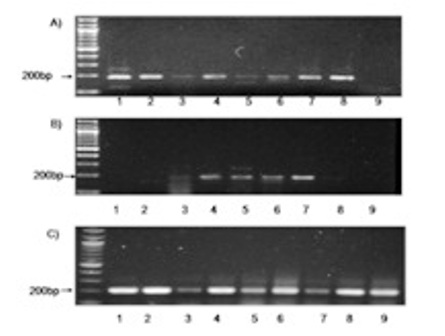
Figure 1: RT PCR analysis of messenger RNA (mRNA) using primers specific for A) survivin in breast cancer cases (1-9) B) survivin in fibroadenoma cases(1-9) C) Amplification of GAPDH was serially analysed for all samples.
Table 2: Correlation between clinico-pathological factors and expression of survivin at mRNA level in breast cancer cases.
| Variable |
Cancer(RNA) |
Chi
square
(RNA) |
Pvalue
(RNA) |
Menopause
Status
Premenopausal
Postmenopausal |
76.7%(23/30)
70.5%(43/61) |
0.385 |
0.535
ns |
Stage
1+2
3
4 |
50%(16/32)
92.5%(37/40)
68.4%(13/19) |
16.549 |
0.001 |
Lymph Node
negative
Positive |
57.7%(15/26)
78.5%(51/65) |
4.021td>
| 0.045 |
GraGrade
1
2
3 |
55.6%
(10/18)
88.1%(37/42)
61.3%(19/31) |
9.675d>
| 0.008 |
EstEstrogen
Receptor
Negative
positive |
82.3%(51/62)
51.2%(15/29) |
9.245d>
| 0.002 |
ProgProgesterone
Receptor
negative
positive |
78.1%(50/64)
59.3%(16/27) |
3.392>
| 0.066 ns |
| HER2 HER2
positive |
55%(22/40)
86.3%(44/51) |
11.004
| 0.001 |
Blood Blood Group
B+
A+
O+ |
80.6%(29/36)
69.2%(9/13)
66.7%(28/42) |
1.959
| 0.375 0.375
ns |
Statistical analysis
The statistical analysis was performed using SPSS 16 software. Survivin expression and its correlation with other clinico-pathological parameters was studied by chi square statistics, a value of p<0.05 was considered significant.
Table 3: Localisation of survivin expression
|
Cytoplasmic
Expression |
Nuclear
Expression |
Cytoplasmic
and Nuclear
Expression |
Breast
Cancer |
67.2%(39/58) |
19%(11/58) |
13.8%(8/58) |
Fibro-
adenoma |
66.7%(8/12) |
33.3%(4/12) |
0%(0/16) |
Results
Survivin mRNA analysis
Survivin mRNA was expressed in 44.8% (13/29) of fibroadenoma (Table 1,Figure 1A) and 72.5%(66/91) of breast cancers (Table 1,Figure 1) cases. Survivin mRNA expression was found to be higher in breast cancer compared to controls, however the difference in expression was not significant (p=0.064) (Table 1).
Survivin mRNA expression was correlated with clinico-pathological parameters (Table 2). Survivin mRNA expression correlated significantly with tumor stage, histological grade, lymph node, estrogen receptor (ER) and HER 2 status. (Table 2)
Survivin protein analysis
Survivin protein was expressed in 41.3% (12/29) of fibroadenoma and 63.7% (58/91) of breast cancers (Table 1). Immunohistochemical analysis of the patients with breast cancer showed the antigen expression in nucleus (19.0%), cytoplasm (67.2%) and both (13.8%) (Table 3,Figure 2). In fibroadenoma antigen expression was seen in nucleus (33.3%) and cytoplasm (66.7%) (Table 3,Fig 3).Weak to strong survivin expression was observed in both breast cancer cases and controls (Table 4). Survivin expression was found to be higher in breast cancer compared to controls, however the difference in expression was not significant (p=0.079) (Table 1). Survivin protein was not found in any sample lacking detectable survivin mRNA.
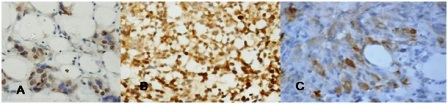
Figure 2: Immunohistochemistry for survivin in breast cancer. a) Nuclear stain positivity; b) Nuclear and cytoplasmic; c) Cytoplasmic stain positivity
Survivin protein was also measured by Western blotting in 20 fibroadenoma and 71 breast cancer cases. Survivin protein was detected in 40% (8/20) of fibroadenoma(Table 1,Fig 4 A) and 64.8% (46/71) of breast cancer cases(Table 1,Fig 4). Survivin expression was higher in breast cancer compared to controls and the difference in expression was significant (p=0.005).(Table 1)
Survivin protein expression showed significant correlation with tumor stage, histological grade, ER, PR and HER2 status (Table 5). Survivin protein expression did not correlate significantly with menstrual status, lymph node and blood group of breast cancer patients (Table 5).
Discussion
Survivin a member of IAP family, located on chromosome 17q25,comprises three introns and four exons, encoding 142 amino acids, including a single BIR domain which is essential for apoptosis inhibition. Survivin or Baculoviral inhibitor of apoptosis repeat-containing protein 5 (BIRC5) is present at low or undetectable levels in normal tissues, where it regulates mitosis and prevents caspase activation [19,20]. It is one of the most uniformly upregulated genes in tumor tissues, necessary for extension of the limited cellular lifespan in cancer via inhibition of apoptosis [21].Our results showed survivin mRNA expression to be higher in breast cancer cases as compared to fibroadenoma ,these findings are in agreement with that of Ryan et al.[22]. Survivin has been described both as a chromosomal passenger protein in the nucleus and as a cytoplasmic microtubule associated protein. Our results of survivin protein expression by Immunohistochemistry method was similar to Al-Joudi F S et al,[23]. Survivin protein expression in both fibroadenoma and breast cancer cases by western blotting showed almost similar expression as Immunohistochemistry. Immunohistochemical analysis of protein showed expression in nucleus (19%), cytoplasm (67.2%), while 13.8% showed both nuclear and cytoplasmic expression. This study shows that survivin is predominantly expressed in cytoplasm in agreement with previous studies [24,25,26], where caspases are present and hence can be said to be effective in blocking apoptosis. An earlier study has suggested that cytosolic pool is associated with interphase microtubule, centrosomes, spindle pools and mitotic spindle microtubules at metaphase and anaphase[27]. It has also been stated that only cytosolic survivin associates with p34cdc2 and phosphorylation of survivin by p34cdc2-cyclinB has been identified as a requisite for apoptosis inhibition [28]. Hence, cytoplasmic protein can be used as a useful biomarker in diagnosis.
Table 4:Survivin: Immunostaining Intensity
|
Weak(+) |
Moderate(++) |
Strong(+++) |
| Breast Cancer |
63.7%(37/58) |
34.4%(20/58) |
1.7%(1/58) |
| Fibroadenoma |
58.3%(7/12) |
33.3%(4/12) |
8.3%(1/12) |
Previous results have shown no significant correlation of survivin with clinico-pathological parameters [12,13,23].However, the present results show survivin expression to be significantly correlated with tumor stage, histological grade[10,29], ER and HER2 [10] status (Table 3 and Table 4) at both mRNA and protein level.
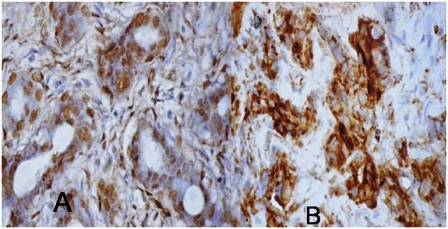
Figure 3: survivin immunohistochemistry in fibroadenoma: A) Ductal cells have intense positivity(Nuclear 3+), B) Cytoplasmic (3+) positivity
Table 5: Correlation between clinicopathological factors and expression of survivin at protein level in breast cancer cases.
| Variable |
Cancer(IHC) |
chi
square
(IHC) |
p value
(IHC) |
Menopause
Status
Pre
Post |
60%(18/30)
65.6%(40/61) |
4.795 |
00.309
ns |
Stage
1+2
3
4 |
40.6%(13/32)
87.5%(35/40)
52.6%(10/19) |
37.252/td>
| 0.000 |
LLymph Node
negative
positive |
50%(13/26)
69.2%(45/65) |
3.617/td>
| 0.0.460
ns |
Grade
1
2
3 |
44.4%(8/18)
80.9%(34/42)
51.6%(16/31) |
19.203td>
| 0.014 |
EsEstrogen
Receptor
Negative
positive |
75.8%(47/62)
37.9%(11/29) |
16.197td>
| 0.003 |
ProProgesterone
Receptor
negative
positive |
71.8%(46/64)
44.4%(12/27) |
12.485d>
| 0.014 |
HER2HER2
Negative
positive |
45.0%(18/40)
78.4%(40/51) |
11.908>
| 0.018 |
BloodBlood Group
B+
A+
O+ |
72.2%(26/36)
69.2%(9/13)
54.8%(23/42) |
6.633
| 0.5770.577
ns |
Survivin expression did not correlate significantly with PR status and correlated significantly with lymph node metastasis at mRNA level. Survivin expression at both mRNA and protein level did not correlate significantly with menstrual status and blood group of breast cancer patients.
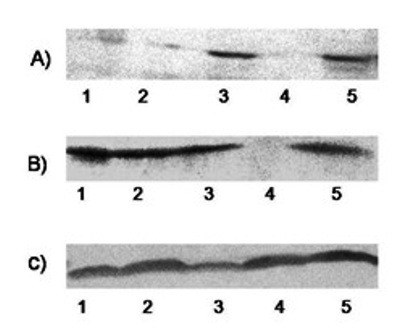
Figure 4: Expression of Survivin protein analyzed using western blotting: A) Fibroadenoma and B) Breast cancer C) beta Actin as control
Conclusions
Our results show that survivin gene and protein is more expressed in breast cancer when compared to fibroadenoma cases. ER negative patients showed greater expression of survivin gene. Increased survivin expression at gene level might serve as a diagnostic and prognostic marker in breast cancer. This gene can be targeted by designing antisense oligonucleotides which binds to survivin RNA to inhibit its function and hence can be used for therapeutic purpose. Further studies on different splice variants of survivin are needed to and establish the biological role for different forms of survivin by using more robust techniques.
Funding Source
This study is supported by a research grant from Indian Council of Medical Research 5/13/120/09-NCD 3
Authors' contribution
KJ: Conducted the work, collection and analysis of data, preparation of manuscript.
MK: Designed and helped in the pathology part of the study and preparation of manuscript
VKS: Helped with collection of data, interpretation of results and editing of the manuscript
MP: Designed the study, coordinated the study, interpretation of results and preparation of manuscript.
All authors read and approved the manuscript for publication.
Conflict of Interests
Authors declare that there are no conflicts of interests.
Ethical Considerations
The study was approved by the Institute Ethics committee.
References
[1]. Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med 2001; 344(4): 276–85.
[2]. Sasco AJ, Kaaks R, Little RE. Breast cancer: occurrence, risk factors and hormone metabolism. Expert Rev Anticancer Ther 2003; 3(4): 546–62.
[3]. Birnbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol 1994; 68: 2521–2528.
[4]. Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting gene with a zinc finger-like motif. J Virol 1993; 67: 2168–2174.
[5]. Reed JC, Doctor KS, Godzik A. The domains of apoptosis: a genomics perspective [RE9]. Sci STKE 2004; 239, 1–29.
[6]. Ryan BM, Konecny GE, Kahlert S, Wang HJ, Untch M, Meng G, Pegram MD, Podratz KC, Crown J, Slamon DJ, Duffy MJ. Survivin expression in breast cancer predicts clinical outcome and is associated with HER2, VEGF, urokinase plasminogen activator and PAI-1. Ann Oncol 2006; 17: 597-604.
[7]. Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem 1998; 273: 11177-82.
[8]. Adida C, Haioun C, Gaulard P, Lepage E, Morel P, Briere J, Dombret H, Reyes F, Diebold J, Gisselbrecht C, Salles G, Altieri DC, Molina TJ. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood 2000; 96: 1921-25.
[9]. Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol 1999; 113: 1076-81.
[10]. Chu JS, ShewJY, Huang CS. Immunohistochemical analysis of survivin expression in primary breast cancers. J Formos Med Assoc 2004; 103: 925-31.
[11]. O'Driscoll L, Cronin D, Kennedy SM, Purcell R, Linehan R, Glynn S, Larkin A, Scanlon K, McDermott EW, Hill AD, O'Higgins NJ, Parkinson M, Clynes M. Lack of prognostic significance of survivin, survivin-DEx3, survivin-2B, galectin-3, bag-1, bax-a and MRP-1 mRNAs in breast cancer. Cancer Lett 2003; 201: 225-36.
[12]. Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res 2000; 6: 127-34.
[13]. Kennedy SM, O’Driscoll L, Purcell R, Fitz-simons N, McDermott EW, Hill AD, O'Higgins NJ, Parkinson M, Linehan R, Clynes M. Prognostic importance of survivin in breast cancer. Br J Cancer 2003; 88: 1077-83.
[14]. Span PN, Sweep FC, Wiegerinck ET, Tjan-Heijnen VC, Manders P, Beex LV, de Kok JB. Survivin is an independent prognostic marker for risk stratification of breast cancer patients. Clin Chem 2004; 50: 1986 - 93.
[15]. Hinnis AR, Luckett JC, Walker RA. Survivin is an independent predictor of short-term survival in poor prognostic breast cancer patients. Br J Cancer 2007; 96: 639-45.
[16]. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004; 351: 2817-26.
[17]. Tsuji N, Furuse k, Asanuma K, Furuya M, Kondoh K, Kamagata C, Sasaki M, Kobayashi D, Yagihashi A, Takahashi H, Watanabe N. Mutations of the p53 gene and loss of heterozygosity at chromosome 17p13.1 are associated with increased survivin expression in breast cancer. Breast Cancer Res Treat 2004; 87: 23–31.
[18]. Xiong H, Yu S, Zhuang L, Xiong H. Changes of survivin mRNA and protein expression during paclitaxel treatment in breast cancer cells. J Huazhong Univ Sci Technolog Med Sci 2007; 27: 65-67.
[19]. Ambrosini G, Adida C, Altieri DC. A novel antiapoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997; 3:917–921.
[20]. Reed JC, Bischoff JR. BIRinging chromosomes through cell division-and survivin’ the experience. Cell 2000;102:545–548.
[21]. Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, Cook BP, Dufault MR, Ferguson AT, Gao Y, He TC, Hermeking H, Hiraldo SK, Hwang PM, Lopez MA, Luderer HF, Mathews B, Petroziello JM, Polyak K, Zawel L, Kinzler KW, et al . Analysis of human transcriptomes. Nat Genet 1999;23:387–388.
[22]. Ryan B, O’Donovan N, Browne B, O’Shea C, Crown J, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Expression of survivin and its splice variants survivin-2B and survivin-DeltaEx3 in breast cancer. Br J Cancer 2005; 92: 120-4.
[23]. Al-Joudi FS, Iskandar ZA, Hasnan J, Rusli J, Kamal Y, Imran AK, Ahmed M, Zakaria J. Expression of survivin and its clinicopathological correlations in invasive ductal carcinoma of the breast. Singapore Med J 2007; 48 (7): 607.
[24]. Okada E, Murai Y, Matsui K, Isizawa S, Cheng C, Masuda M, Takano Y. Survivin expression in tumor cell nuclei is predictive of a favorable prognosis in gastric cancer patients. Cancer Lett 2001; 163: 109–16.
[25]. Dogu GG, Ozkan M, Ozturk F, Dikilitas M, Ozlem Er, Ozturk A. Triple-negative breast cancer: immunohistochemical correlation with basaloid markers and prognostic value of survivin. Med Oncol 2010; 27:34-39.
[26]. Sohn DM, Kim SY, Baek MJ, Lim CW, Lee MH, Cho MS, Kim TY. Expression of survivin and clinical correlation in patients with breast cancer. Biomed Pharmacother 2006; 60: 289-92.
[27]. Fortugno P, Wall NR, Giodini A, O’Connor DS, Plescia J, Padgett KM, Tognin S, Marchisio PC, Altieri DC. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci 2002; 115: 575–585.
[28]. O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC.Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA 2000; 97: 13103–13107.
[29]. Kostadima L, Pentheroudakis G, Fountzilas G, Dimopoulos M, Pectasides D, Gogas H, Stropp U, Christodoulou C, Samantas E, Wirtz R, Hennig G, Bafaloukos D, Arapantoni P, Kalofonos H, Papakostas P, Economopoulos T, Bamias A, Pavlidis N. Survivin and glycodelin transcriptional activity in node-positive early breast cancer: mRNA expression of two key regulators of cell survival .Breast Cancer Res Treat 2006; 100: 161–167.

